Spectral line catalog data. More...
#include <linerecord.h>

Public Types | |
| enum | { BROAD_SPEC_POS_N2, BROAD_SPEC_POS_O2, BROAD_SPEC_POS_H2O, BROAD_SPEC_POS_CO2, BROAD_SPEC_POS_H2, BROAD_SPEC_POS_He } |
| Indices of different broadening species in Gamma_foreign, N_foreign, and Delta_foreign. More... | |
Public Member Functions | |
| LineRecord () | |
| Default constructor. More... | |
| LineRecord (Index species, Index isotopologue, Numeric f, Numeric psf, Numeric i0, Numeric ti0, Numeric elow, Numeric agam, Numeric sgam, Numeric nair, Numeric nself, Numeric tgam, const ArrayOfNumeric &aux, Numeric, Numeric, Numeric, Numeric, Numeric, Numeric, Numeric) | |
| Constructor that sets all data elements explicitly. More... | |
| String | VersionString () const |
| Return the version String. More... | |
| Index | Version () const |
| Return the version number. More... | |
| Index | Species () const |
| The index of the molecular species that this line belongs to. More... | |
| Index | Isotopologue () const |
| The index of the isotopologue species that this line belongs to. More... | |
| String | Name () const |
| The full name of the species and isotopologue. More... | |
| const SpeciesRecord & | SpeciesData () const |
| The matching SpeciesRecord from species_data. More... | |
| const IsotopologueRecord & | IsotopologueData () const |
| The matching IsotopologueRecord from species_data. More... | |
| Numeric | F () const |
| The line center frequency in Hz. More... | |
| void | setF (Numeric new_mf) |
| Set the line center frequency in Hz. More... | |
| Numeric | Psf () const |
| The pressure shift parameter in Hz/Pa. More... | |
| void | setPsf (Numeric new_mpsf) |
| Set the pressure shift parameter in Hz/Pa. More... | |
| Numeric | I0 () const |
The line intensity in m^2*Hz at the reference temperature Ti0. More... | |
| void | setI0 (Numeric new_mi0) |
| Set Intensity. More... | |
| Numeric | Ti0 () const |
| Reference temperature for I0 in K: More... | |
| Numeric | Elow () const |
| Lower state energy in cm^-1: More... | |
| Numeric | Agam () const |
| Air broadened width in Hz/Pa: More... | |
| void | setAgam (Numeric new_agam) |
| Set Air broadened width in Hz/Pa: More... | |
| Numeric | Sgam () const |
| Self broadened width in Hz/Pa: More... | |
| void | setSgam (Numeric new_sgam) |
| Set Self broadened width in Hz/Pa: More... | |
| Numeric | Nair () const |
| AGAM temperature exponent (dimensionless): More... | |
| void | setNair (Numeric new_mnair) |
| Set AGAM temperature exponent (dimensionless): More... | |
| Numeric | Nself () const |
| SGAM temperature exponent (dimensionless): More... | |
| void | setNself (Numeric new_mnself) |
| Set SGAM temperature exponent (dimensionless): More... | |
| Numeric | Tgam () const |
| Reference temperature for AGAM and SGAM in K: More... | |
| Index | Naux () const |
| Number of auxiliary parameters. More... | |
| const ArrayOfNumeric & | Aux () const |
| Auxiliary parameters. More... | |
| Numeric | dF () const |
| Accuracy for line position in Hz : More... | |
| Numeric | dI0 () const |
| Accuracy for line intensity in relative value : More... | |
| Numeric | dAgam () const |
| Accuracy for air broadened width in relative value : More... | |
| Numeric | dSgam () const |
| Accuracy for self broadened width in relative value : More... | |
| Numeric | dNair () const |
| Accuracy for AGAM temperature exponent in relative value : More... | |
| Numeric | dNself () const |
| Accuracy for SGAM temperature exponent in relative value: More... | |
| Numeric | dPsf () const |
| Accuracy for pressure shift in relative value : More... | |
| Numeric | A () const |
| ARTSCAT-4 Einstein A-coefficient in 1/s : More... | |
| Numeric | G_upper () const |
| ARTSCAT-4 Upper state stat. More... | |
| Numeric | G_lower () const |
| ARTSCAT-4 Lower state stat. More... | |
| Numeric | Gamma_foreign (const Index i) const |
| ARTSCAT-4 foreign broadening parameters in Hz/Pa : More... | |
| Numeric | N_foreign (const Index i) const |
| ARTSCAT-4 foreign temperature exponents (dimensionless): More... | |
| Numeric | Delta_foreign (const Index i) const |
| ARTSCAT-4 pressure shift parameters in Hz/Pa : More... | |
| const String & | Upper_GQuanta () const |
| Broadening parameter self in Hz/Pa : More... | |
| const String & | Lower_GQuanta () const |
| Lower state global quanta. More... | |
| const String & | Upper_LQuanta () const |
| Upper state local quanta. More... | |
| const String & | Lower_LQuanta () const |
| Lower state local quanta. More... | |
| Rational | Upper_N () const |
| Upper state local quanta N. More... | |
| Rational | Upper_J () const |
| Upper state local quanta J. More... | |
| Rational | Lower_N () const |
| Lower state local quanta N. More... | |
| Rational | Lower_J () const |
| Lower state local quanta J. More... | |
| const String & | QuantumNumbersString () const |
| String with quantum numbers. More... | |
| const QuantumNumberRecord & | QuantumNumbers () const |
| Quantum numbers. More... | |
| void | ARTSCAT4FromARTSCAT3 () |
| Converts line parameters from ARTSCAT-3 to ARTSCAT-4 format. More... | |
| void | ARTSCAT4UnusedToNaN () |
| Set to NaN all parameters that are not in ARTSCAT-4. More... | |
| bool | ReadFromHitran2001Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with a HITRAN 1986-2001 file. More... | |
| bool | ReadFromHitran2004Stream (istream &is, const Verbosity &verbosity, const Numeric fmin=0) |
| Read one line from a stream associated with a HITRAN 2004 file. More... | |
| bool | ReadFromMytran2Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with a MYTRAN2 file. More... | |
| bool | ReadFromJplStream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with a JPL file. More... | |
| bool | ReadFromArtscat3Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with an ARTSCAT-3 file. More... | |
| bool | ReadFromArtscat4Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with an ARTSCAT-4 file. More... | |
Static Public Member Functions | |
| static Index | NBroadSpec () |
| Return the number of artscat-4 foreign broadening species (6). More... | |
| static String | BroadSpecName (const Index i) |
| Return the name of an artscat-4 broadening species, as function of its broadening species index. More... | |
| static Index | BroadSpecSpecIndex (const Index i) |
| Return the internal species index (index in species_data) of an artscat-4 broadening species, as function of its broadening spcecies index. More... | |
Detailed Description
Spectral line catalog data.
Below is a description of the ARTS catalogue format. The file starts with the usual XML header:
<?xml version="1.0"?>
<arts format="ascii" version="1">
<ArrayOfLineRecord version="ARTSCAT-3" nelem="8073">The ARTSCAT version number is there to keep track of catalogue format changes. The "nelem" tag contains the total number of lines in the file.
The file ends with the usual XML closing tags:
</ArrayOfLineRecord>
</arts>In-between the header and the footer are the actual spectroscopic data. Each new entry, corresponding to one spectral line, starts with the `@' character.
The line catalogue should not have any fixed column widths because the precision of the parameters should not be limited by the format. The catalogue can then be stored as binary or ASCII. In the ASCII version the columns are separated by one or more blanks. The line format is then specified by only the order and the units of the columns. As the catalogue entry for each transition can be quite long, it can be broken across lines in the ASCII file. That is why each new transition is marked with a `@' character.
The first column will contain the species and isotopologue, following the naming scheme described below. Scientific notation is allowed, e.g. 501.12345e9.
Note that starting with ARTSCAT-2, the intensity is per molecule, i.e., it does not contain the isotopologue ratio. This is similar to JPL, but different to HITRAN.
Currently, ARTS is capable of handling ARTSCAT versions 3 and 4. Different versions can be handled simultaneously. The line format of ARTSCAT-3 (for ARTSCAT-4 see further below) is:
Col Variable Label Unit Comment
------------------------------------------------------------------
0 `@' ENTRY - marks start of entry
1 species\&isotopologue tag NAME - e.g. O3-666
2 center frequency F Hz e.g. 501.12345e9
3 pressure shift of F PSF Hz/Pa
4 line intensity I0 Hz*m^2 per isotopologue, not per species
5 reference temp. for I0 T_I0 K
6 lower state energy ELOW J
7 air broadened width AGAM Hz/Pa values around 20000 Hz/Pa
8 self broadened width SGAM Hz/Pa
9 AGAM temp. exponent NAIR - values around .5
10 SGAM temp. exponent NSELF -
11 ref. temp. for AGAM, SGAM T_GAM K
12 number of aux. parameters N_AUX -
13 auxiliary parameter AUX1 -
14 ...
15 error for F DF Hz
16 error for I0 DI0 %
17 error for AGAM DAGAM %
18 error for SGAM DSGAM %
19 error for NAIR DNAIR %
20 error for NSELF DNSELF %
21 error for PSF DPSF %The parameters 0-12 must be present, the others can be missing, since they are not needed for the calculation.
For the error fields (15-21), a -1 means that no value exist.
A valid ARTS (CAT-3) line file would be:
<?xml version="1.0"?>
<arts format="ascii" version="1">
<ArrayOfLineRecord version="ARTSCAT-3" nelem="2">
@ O3-676 80015326542.0992 0 3.70209114155527e-19 296 7.73661776567701e-21 21480.3182341969
28906.7092490501 0.76 0.76 296 0 300000 0.1 0.1 0.1 0.2 -1 -1.24976056865038e-11
@ O3-676 80015476438.3282 0 3.83245786810611e-19 296 7.73661577919822e-21 21480.3182341969
28906.7092490501 0.76 0.76 296 0 300000 0.1 0.1 0.1 0.2 -1 -1.24975822742335e-11
</ArrayOfLineRecord>
</arts>Some species need special parameters that are not needed by other species (for example overlap coefficients for O2). In the case of oxygen two parameters are sufficient to describe the overlap, but other species, e.g., methane, may need more coefficients. The default for N_AUX is zero. In that case, no further AUX fields are present. [FIXME: Check Oxygen.]
The line format of ARTSCAT-4 is:
Col Variable Label Unit Comment
------------------------------------------------------------------
00 `@' ENTRY - marks start of entry
01 species\&isotopologue tag NAME - e.g. O3-666
02 center frequency F Hz e.g. 501.12345e9
03 line intensity I0 Hz*m^2 per isotopologue, not per species
04 reference temp. for I0 T_I0 K
05 lower state energy ELOW J
06 Einstein A-coefficient A 1/s where available from HITRAN
07 Upper state stat. weight G_upper - where available from HITRAN
08 Lower state stat. weight G_lower - where available from HITRAN
09 broadening parameter self GAMMA_self Hz/Pa GAM have values around 20000 Hz/Pa
10 broadening parameter N2 GAMMA_N2 Hz/Pa
11 broadening parameter O2 GAMMA_O2 Hz/Pa
12 broadening parameter H2O GAMMA_H2O Hz/Pa
13 broadening parameter CO2 GAMMA_CO2 Hz/Pa
14 broadening parameter H2 GAMMA_H2 Hz/Pa
15 broadening parameter He GAMMA_He Hz/Pa
16 GAM temp. exponent self N_self - N have values around .5
17 GAM temp. exponent N2 N_N2 -
18 GAM temp. exponent O2 N_O2 -
19 GAM temp. exponent H2O N_H2O -
20 GAM temp. exponent CO2 N_CO2 -
21 GAM temp. exponent H2 N_H2 -
22 GAM temp. exponent He N_He -
23 F pressure shift N2 DELTA_N2 Hz/Pa DELTA have values around 0 Hz/Pa
24 F pressure shift O2 DELTA_O2 Hz/Pa
25 F pressure shift H2O DELTA_H2O Hz/Pa
26 F pressure shift CO2 DELTA_CO2 Hz/Pa
27 F pressure shift H2 DELTA_H2 Hz/Pa
28 F pressure shift He DELTA_He Hz/Pa
29 Vibrational and rotational VRA - contains (coded) quantum numbers.
assignments
------------------------------------------------------------------
Parameters 0-28 must be present. Coding conventions of parameter 29 are species specific. The definition is given in arts-xml-data/spectroscopy/perrin/ARTSCAT-4_Col29_Conventions.txt. The names of the private members and public access functions of this data structure follow the above table. The only difference is that underscores are omited and only the first letter of each name is capitalized. This is for consistency with the notation elsewhere in the program. \author Stefan Buehler
Definition at line 196 of file linerecord.h.
Member Enumeration Documentation
◆ anonymous enum
| anonymous enum |
Indices of different broadening species in Gamma_foreign, N_foreign, and Delta_foreign.
| Enumerator | |
|---|---|
| BROAD_SPEC_POS_N2 | |
| BROAD_SPEC_POS_O2 | |
| BROAD_SPEC_POS_H2O | |
| BROAD_SPEC_POS_CO2 | |
| BROAD_SPEC_POS_H2 | |
| BROAD_SPEC_POS_He | |
Definition at line 543 of file linerecord.h.
Constructor & Destructor Documentation
◆ LineRecord() [1/2]
|
inline |
Default constructor.
Initialize to default values. The indices are initialized to large numbers, so that we at least get range errors when we try to used un-initialized data.
Definition at line 202 of file linerecord.h.
◆ LineRecord() [2/2]
|
inline |
Constructor that sets all data elements explicitly.
If assertions are not disabled (i.e., if NDEBUG is not #defined), assert statements check that the species and isotopologue data exists.
Definition at line 236 of file linerecord.h.
References VersionString().
Member Function Documentation
◆ A()
|
inline |
ARTSCAT-4 Einstein A-coefficient in 1/s :
Definition at line 433 of file linerecord.h.
References ma.
Referenced by operator<<().
◆ Agam()
|
inline |
Air broadened width in Hz/Pa:
Definition at line 374 of file linerecord.h.
References magam.
Referenced by operator<<(), and xsec_species().
◆ ARTSCAT4FromARTSCAT3()
| void LineRecord::ARTSCAT4FromARTSCAT3 | ( | ) |
Converts line parameters from ARTSCAT-3 to ARTSCAT-4 format.
ARTSCAT-4 lines contain more information than ARTSCAT-3 lines, particularly they contain separate broadening parameters for six different broadening species. So a real conversion is not possible. What this method does is copy the air broadening (and shift) parameters from ARTSCAT-3 to all ARTSCAT-4 broadening species. The case that one of the broadening species is identical to the Self species is also handled correctly.
The idea is that the ARTSCAT-4 line list generated in this way should give identical RT simulation results as the original ARTSCAT-3 list. This is verified in one of the test controlfiles.
Currently only broadening and shift parameters are handled here. There are some other additional fields in ARTSCAT-4, which we so far ignore.
Definition at line 71 of file linerecord.cc.
References ARTSCAT4UnusedToNaN(), BroadSpecSpecIndex(), magam, mdelta_foreign, mgamma_foreign, mn_foreign, mnair, mnself, mpsf, msgam, mversion, NBroadSpec(), Vector::resize(), Species(), and Version().
Referenced by BroadSpecName().
◆ ARTSCAT4UnusedToNaN()
|
inline |
Set to NaN all parameters that are not in ARTSCAT-4.
Definition at line 612 of file linerecord.h.
References magam, maux, mdagam, mdf, mdi0, mdnair, mdnself, mdpsf, mdsgam, mnair, mpsf, mtgam, ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), and ReadFromMytran2Stream().
Referenced by ARTSCAT4FromARTSCAT3(), and ReadFromArtscat4Stream().
◆ Aux()
|
inline |
Auxiliary parameters.
Definition at line 408 of file linerecord.h.
References maux.
Referenced by operator<<().
◆ BroadSpecName()
Return the name of an artscat-4 broadening species, as function of its broadening species index.
Meant to be called with the enum constants defined in this class.
Definition at line 559 of file linerecord.h.
References ARTSCAT4FromARTSCAT3(), BROAD_SPEC_POS_CO2, BROAD_SPEC_POS_H2, BROAD_SPEC_POS_H2O, BROAD_SPEC_POS_He, BROAD_SPEC_POS_N2, BROAD_SPEC_POS_O2, and BroadSpecSpecIndex().
Referenced by BroadSpecSpecIndex(), and calc_gamma_and_deltaf_artscat4().
◆ BroadSpecSpecIndex()
Return the internal species index (index in species_data) of an artscat-4 broadening species, as function of its broadening spcecies index.
Meant to be called with the enum constants defined in this class.
Definition at line 64 of file linerecord.cc.
References BroadSpecName(), and species_index_from_species_name().
Referenced by ARTSCAT4FromARTSCAT3(), BroadSpecName(), and find_broad_spec_locations().
◆ dAgam()
|
inline |
Accuracy for air broadened width in relative value :
Definition at line 418 of file linerecord.h.
References mdagam.
Referenced by operator<<().
◆ Delta_foreign()
ARTSCAT-4 pressure shift parameters in Hz/Pa :
Definition at line 448 of file linerecord.h.
References mdelta_foreign.
Referenced by calc_gamma_and_deltaf_artscat4(), and operator<<().
◆ dF()
|
inline |
Accuracy for line position in Hz :
Definition at line 412 of file linerecord.h.
References mdf.
Referenced by operator<<().
◆ dI0()
|
inline |
Accuracy for line intensity in relative value :
Definition at line 415 of file linerecord.h.
References mdi0.
Referenced by operator<<().
◆ dNair()
|
inline |
Accuracy for AGAM temperature exponent in relative value :
Definition at line 424 of file linerecord.h.
References mdnair.
Referenced by operator<<().
◆ dNself()
|
inline |
Accuracy for SGAM temperature exponent in relative value:
Definition at line 427 of file linerecord.h.
References mdnself.
Referenced by operator<<().
◆ dPsf()
|
inline |
Accuracy for pressure shift in relative value :
Definition at line 430 of file linerecord.h.
References mdpsf.
Referenced by operator<<().
◆ dSgam()
|
inline |
Accuracy for self broadened width in relative value :
Definition at line 421 of file linerecord.h.
References mdsgam.
Referenced by operator<<().
◆ Elow()
|
inline |
Lower state energy in cm^-1:
Definition at line 371 of file linerecord.h.
References melow.
Referenced by operator<<(), and xsec_species().
◆ F()
|
inline |
The line center frequency in Hz.
Definition at line 339 of file linerecord.h.
References mf.
Referenced by abs_lines_per_speciesAddMirrorLines(), abs_lines_per_speciesCreateFromLines(), abs_linesReadFromArtsObsolete(), abs_linesReadFromHitran(), abs_linesReadFromHitranPre2004(), abs_linesReadFromJpl(), abs_linesReadFromMytran2(), operator<<(), xml_read_from_stream(), and xsec_species().
◆ G_lower()
|
inline |
ARTSCAT-4 Lower state stat.
weight:
Definition at line 439 of file linerecord.h.
References mglower.
Referenced by operator<<().
◆ G_upper()
|
inline |
ARTSCAT-4 Upper state stat.
weight:
Definition at line 436 of file linerecord.h.
References mgupper.
Referenced by operator<<().
◆ Gamma_foreign()
ARTSCAT-4 foreign broadening parameters in Hz/Pa :
Definition at line 442 of file linerecord.h.
References mgamma_foreign.
Referenced by calc_gamma_and_deltaf_artscat4(), and operator<<().
◆ I0()
|
inline |
The line intensity in m^2*Hz at the reference temperature Ti0.
The line intensity 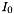 is defined by:
is defined by:
![\[ \alpha(\nu) = n \, x \, I_0(T) \, F(\nu) \]](form_6.png)
where 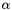 is the absorption coefficient (in m^-1),
is the absorption coefficient (in m^-1), 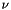 is frequency,
is frequency, 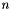 is the total number density,
is the total number density, 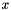 is the volume mixing ratio, and
is the volume mixing ratio, and 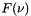 is the lineshape function.
is the lineshape function.
Definition at line 362 of file linerecord.h.
References mi0.
Referenced by operator<<(), and xsec_species().
◆ Isotopologue()
|
inline |
The index of the isotopologue species that this line belongs to.
The isotopologue species data can be accessed by species_data[Species()].Isotopologue()[Isotopologue()].
Definition at line 307 of file linerecord.h.
References IsotopologueData(), misotopologue, Name(), and SpeciesData().
Referenced by abs_lines_per_speciesCreateFromLines(), find_matching_lines(), propmat_clearskyAddZeeman(), and xsec_species().
◆ IsotopologueData()
| const IsotopologueRecord & LineRecord::IsotopologueData | ( | ) | const |
The matching IsotopologueRecord from species_data.
The IsotopologueRecord is a subset of the SpeciesRecord. To get at the isotopologue data of a LineRecord lr, you can use:
- species_data[lr.Species()].Isotopologue()[lr.Isotopologue()]
- lr.SpeciesData().Isotopologue()[lr.Isotopologue()]
- lr.IsotopologueData()
The last option is clearly the shortest, and has the advantage that you don't have to declare the external variable species_data.
Definition at line 57 of file linerecord.cc.
References misotopologue, mspecies, and global_data::species_data.
Referenced by Isotopologue(), and xsec_species().
◆ Lower_GQuanta()
|
inline |
◆ Lower_J()
|
inline |
◆ Lower_LQuanta()
|
inline |
◆ Lower_N()
|
inline |
◆ N_foreign()
ARTSCAT-4 foreign temperature exponents (dimensionless):
Definition at line 445 of file linerecord.h.
References mn_foreign.
Referenced by calc_gamma_and_deltaf_artscat4(), and operator<<().
◆ Nair()
|
inline |
AGAM temperature exponent (dimensionless):
Definition at line 386 of file linerecord.h.
References mnair.
Referenced by operator<<(), and xsec_species().
◆ Name()
| String LineRecord::Name | ( | ) | const |
The full name of the species and isotopologue.
E.g., `O3-666'. The name is found by looking up the information in species_data, using the species and isotopologue index.
Definition at line 42 of file linerecord.cc.
References SpeciesRecord::Isotopologue(), misotopologue, mspecies, SpeciesRecord::Name(), and global_data::species_data.
Referenced by Isotopologue(), operator<<(), ReadFromArtscat4Stream(), and ReadFromHitran2004Stream().
◆ Naux()
|
inline |
Number of auxiliary parameters.
This function is actually redundant, since the number of auxiliary parameters can also be obtained directly with Aux.nelem(). I just added the function in order to have consistency of the interface with the catalgue format.
Definition at line 405 of file linerecord.h.
References maux, and Array< base >::nelem().
Referenced by operator<<().
◆ NBroadSpec()
|
inlinestatic |
Return the number of artscat-4 foreign broadening species (6).
This just so that we do not have to hardwire the number elsewhere.
Definition at line 554 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), calc_gamma_and_deltaf_artscat4(), and find_broad_spec_locations().
◆ Nself()
|
inline |
SGAM temperature exponent (dimensionless):
Definition at line 392 of file linerecord.h.
References mnself.
Referenced by calc_gamma_and_deltaf_artscat4(), operator<<(), and xsec_species().
◆ Psf()
|
inline |
The pressure shift parameter in Hz/Pa.
Definition at line 345 of file linerecord.h.
References mpsf.
Referenced by operator<<(), and xsec_species().
◆ QuantumNumbers()
|
inline |
Quantum numbers.
Definition at line 538 of file linerecord.h.
References mquantum_numbers.
Referenced by find_matching_lines(), and propmat_clearskyAddZeeman().
◆ QuantumNumbersString()
|
inline |
String with quantum numbers.
Definition at line 535 of file linerecord.h.
References mquantum_numbers_str.
Referenced by operator<<().
◆ ReadFromArtscat3Stream()
| bool LineRecord::ReadFromArtscat3Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with an ARTSCAT-3 file.
Format: see Documentation of class LineRecord
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 2000-12-15
- Date
- 2001-06-20
Definition at line 2064 of file linerecord.cc.
References CREATE_OUT3, extract(), SpeciesRecord::Isotopologue(), magam, maux, mdagam, mdf, mdi0, mdnair, mdnself, mdpsf, mdsgam, melow, mf, mi0, misotopologue, mnair, mnself, mpsf, msgam, mspecies, mtgam, mti0, mversion, SpeciesRecord::Name(), my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, and SpecIsoMap::Speciesindex().
Referenced by abs_linesReadFromArtsObsolete(), ARTSCAT4UnusedToNaN(), and xml_read_from_stream().
◆ ReadFromArtscat4Stream()
| bool LineRecord::ReadFromArtscat4Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with an ARTSCAT-4 file.
Format: see Documentation of class LineRecord
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 2012-02-10
Definition at line 2259 of file linerecord.cc.
References ARTSCAT4UnusedToNaN(), CREATE_OUT3, extract(), SpeciesRecord::Isotopologue(), ma, mdelta_foreign, melow, mf, mgamma_foreign, mglower, mgupper, mi0, misotopologue, mlower_j, mlower_n, mn_foreign, mnself, mquantum_numbers, mquantum_numbers_str, msgam, mspecies, mti0, mupper_j, mupper_n, mversion, Name(), SpeciesRecord::Name(), my_basic_string< charT >::nelem(), Array< base >::nelem(), q, QN_F, QN_J, QN_N, QN_v1, QN_v2, QN_v3, Vector::resize(), QuantumNumberRecord::SetLower(), QuantumNumberRecord::SetUpper(), global_data::species_data, SpecIsoMap::Speciesindex(), and my_basic_string< charT >::trim().
Referenced by ARTSCAT4UnusedToNaN(), and xml_read_from_stream().
◆ ReadFromHitran2001Stream()
| bool LineRecord::ReadFromHitran2001Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with a HITRAN 1986-2001 file.
The HITRAN format is as follows (directly from the HITRAN documentation):
Each line consists of 100
bytes of ASCII text data, followed by a line feed (ASCII 10) and
carriage return (ASCII 13) character, for a total of 102 bytes per line.
Each line can be read using the following READ and FORMAT statement pair
(for a FORTRAN sequential access read):
READ(3,800) MO,ISO,V,S,R,AGAM,SGAM,E,N,d,V1,V2,Q1,Q2,IERF,IERS,
* IERH,IREFF,IREFS,IREFH
800 FORMAT(I2,I1,F12.6,1P2E10.3,0P2F5.4,F10.4,F4.2,F8.6,2I3,2A9,3I1,3I2)
Each item is defined below, with its format shown in parenthesis.
MO (I2) = molecule number
ISO (I1) = isotopologue number (1 = most abundant, 2 = second, etc)
V (F12.6) = frequency of transition in wavenumbers (cm-1)
S (E10.3) = intensity in cm-1/(molec * cm-2) at 296 Kelvin
R (E10.3) = transition probability squared in Debyes**2
AGAM (F5.4) = air-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin
SGAM (F5.4) = self-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin
E (F10.4) = lower state energy in wavenumbers (cm-1)
N (F4.2) = coefficient of temperature dependence of air-broadened halfwidth
d (F8.6) = shift of transition due to pressure (cm-1)
V1 (I3) = upper state global quanta index
V2 (I3) = lower state global quanta index
Q1 (A9) = upper state local quanta
Q2 (A9) = lower state local quanta
IERF (I1) = accuracy index for frequency reference
IERS (I1) = accuracy index for intensity reference
IERH (I1) = accuracy index for halfwidth reference
IREFF (I2) = lookup index for frequency
IREFS (I2) = lookup index for intensity
IREFH (I2) = lookup index for halfwidth
The molecule numbers are encoded as shown in the table below:
0= Null 1= H2O 2= CO2 3= O3 4= N2O 5= CO
6= CH4 7= O2 8= NO 9= SO2 10= NO2 11= NH3
12= HNO3 13= OH 14= HF 15= HCl 16= HBr 17= HI
18= ClO 19= OCS 20= H2CO 21= HOCl 22= N2 23= HCN
24=CH3Cl 25= H2O2 26= C2H2 27= C2H6 28= PH3 29= COF2
30= SF6 31= H2S 32=HCOOH
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped. It is checked if the data record has the right number of characters. If not, a runtime_error is thrown.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
Definition at line 116 of file linerecord.cc.
References ATM2PA, convHitranIERF(), convHitranIERSH(), CREATE_OUT0, CREATE_OUT3, extract(), iso(), SpeciesRecord::Isotopologue(), magam, max, mdagam, mdf, mdi0, mdpsf, melow, mf, mi0, misotopologue, mnair, mnself, mpsf, msgam, mspecies, mtgam, mti0, my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, SPEED_OF_LIGHT, and wavenumber_to_joule().
Referenced by abs_linesReadFromHitranPre2004(), and ARTSCAT4UnusedToNaN().
◆ ReadFromHitran2004Stream()
| bool LineRecord::ReadFromHitran2004Stream | ( | istream & | is, |
| const Verbosity & | verbosity, | ||
| const Numeric | fmin = 0 |
||
| ) |
Read one line from a stream associated with a HITRAN 2004 file.
The HITRAN format is as follows:
Each line consists of 160 ASCII characters, followed by a line feed (ASCII 10) and carriage return (ASCII 13) character, for a total of 162 bytes per line. Each item is defined below, with its Fortran format shown in parenthesis. (I2) molecule number (I1) isotopologue number (1 = most abundant, 2 = second, etc) (F12.6) vacuum wavenumbers (cm-1) (E10.3) intensity in cm-1/(molec * cm-2) at 296 Kelvin (E10.3) Einstein-A coefficient (s-1) (F5.4) air-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin (F5.4) self-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin (F10.4) lower state energy (cm-1) (F4.2) coefficient of temperature dependence of air-broadened halfwidth (F8.6) air-broadened pressure shift of line transition at 296 K (cm-1) (A15) upper state global quanta (A15) lower state global quanta (A15) upper state local quanta (A15) lower state local quanta (I1) uncertainty index for wavenumber (I1) uncertainty index for intensity (I1) uncertainty index for air-broadened half-width (I1) uncertainty index for self-broadened half-width (I1) uncertainty index for temperature dependence (I1) uncertainty index for pressure shift (I2) index for table of references correspond. to wavenumber (I2) index for table of references correspond. to intensity (I2) index for table of references correspond. to air-broadened half-width (I2) index for table of references correspond. to self-broadened half-width (I2) index for table of references correspond. to temperature dependence (I2) index for table of references correspond. to pressure shift (A1) flag (*) for lines supplied with line-coupling algorithm (F7.1) upper state statistical weight (F7.1) lower state statistical weight The molecule numbers are encoded as shown in the table below: 0= Null 1= H2O 2= CO2 3= O3 4= N2O 5= CO 6= CH4 7= O2 8= NO 9= SO2 10= NO2 11= NH3 12= HNO3 13= OH 14= HF 15= HCl 16= HBr 17= HI 18= ClO 19= OCS 20= H2CO 21= HOCl 22= N2 23= HCN 24=CH3Cl 25= H2O2 26= C2H2 27= C2H6 28= PH3 29= COF2 30= SF6 31= H2S 32=HCOOH 33= HO2 34= O 35=ClONO2 36= NO+ 37= HOBr 38= C2H4
CH3OH is not included in ARTS because its total internal partition sum is not known yet.
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped. It is checked if the data record has the right number of characters (comment lines are ignored). If not, a runtime_error is thrown. If the molecule is unknown to ARTS, a warning is prompted but the program continues (ignoring this record). For CH3OH this warning will be issued even when using the regular Hitran 2004 data base (see above). If the line center is below fmin, mf is set to -1 and the caller should ignore this line.
- Parameters
-
is Stream from which to read verbosity Verbosity fmin Skip line if mf < fmin
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
Definition at line 558 of file linerecord.cc.
References ATM2PA, convHitranIERF(), convHitranIERSH(), CREATE_OUT1, CREATE_OUT3, extract(), iso(), SpeciesRecord::Isotopologue(), QuantumNumberRecord::Lower(), magam, max, mdagam, mdf, mdi0, mdnair, mdnself, mdpsf, mdsgam, melow, mf, mi0, misotopologue, mlower_gquanta, mlower_j, mlower_lquanta, mlower_n, mnair, mnself, mpsf, mquantum_numbers, msgam, mspecies, mtgam, mti0, mupper_gquanta, mupper_j, mupper_lquanta, mupper_n, Name(), my_basic_string< charT >::nelem(), Array< base >::nelem(), QN_F, QN_J, QN_K1, QN_K2, QN_N, QN_Omega, QN_v1, QuantumNumberRecord::SetLower(), QuantumNumberRecord::SetUpper(), global_data::species_data, SPEED_OF_LIGHT, my_basic_string< charT >::split(), my_basic_string< charT >::trim(), QuantumNumberRecord::Upper(), and wavenumber_to_joule().
Referenced by abs_linesReadFromHitran(), and ARTSCAT4UnusedToNaN().
◆ ReadFromJplStream()
| bool LineRecord::ReadFromJplStream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with a JPL file.
The JPL format is as follows (directly taken from the jpl documentation):
The catalog line files are composed of 80-character lines, with one
line entry per spectral line. The format of each line is:
\label{lfmt}
\begin{tabular}{@{}lccccccccr@{}}
FREQ, & ERR, & LGINT, & DR, & ELO, & GUP, & TAG, & QNFMT, & QN${'}$, & QN${''}$\\
(F13.4, & F8.4, & F8.4, & I2, & F10.4, & I3, & I7, & I4, & 6I2, & 6I2)\\
\end{tabular}
\begin{tabular}{lp{4.5in}}
FREQ: & Frequency of the line in MHz.\\
ERR: & Estimated or experimental error of FREQ in MHz.\\
LGINT: &Base 10 logarithm of the integrated intensity
in units of \linebreak nm$^2$$\cdot$MHz at 300 K. (See Section 3 for
conversions to other units.)\\
DR: & Degrees of freedom in the rotational partition
function (0 for atoms, 2 for linear molecules, and 3 for nonlinear
molecules).\\
ELO: &Lower state energy in cm$^{-1}$ relative to the lowest energy
spin--rotation level in ground vibronic state.\\
GUP: & Upper state degeneracy.\\
TAG: & Species tag or molecular identifier.
A negative value flags that the line frequency has
been measured in the laboratory. The absolute value of TAG is then the
species tag and ERR is the reported experimental error. The three most
significant digits of the species tag are coded as the mass number of the
species, as explained above.\\
QNFMT: &Identifies the format of the quantum numbers
given in the field QN. These quantum number formats are given in Section 5
and are different from those in the first two editions of the catalog.\\
QN${'}$: & Quantum numbers for the upper state coded
according to QNFMT.\\
QN${''}$: & Quantum numbers for the lower state.\\
\end{tabular}
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped (unused in jpl).
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 01.11.00
Definition at line 1818 of file linerecord.cc.
References CREATE_OUT3, extract(), SpeciesRecord::Isotopologue(), magam, mdf, melow, mf, mi0, misotopologue, mnair, mnself, mpsf, msgam, mspecies, mtgam, mti0, my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, SpecIsoMap::Speciesindex(), and wavenumber_to_joule().
Referenced by abs_linesReadFromJpl(), and ARTSCAT4UnusedToNaN().
◆ ReadFromMytran2Stream()
| bool LineRecord::ReadFromMytran2Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with a MYTRAN2 file.
The MYTRAN2 format is as follows (directly taken from the abs_my.c documentation):
The MYTRAN format is as follows (FORTRAN notation):
FORMAT(I2,I1,F13.4,1PE10.3,0P2F5.2,F10.4,2F4.2,F8.6,F6.4,2I3,2A9,4I1,3I2)
Each item is defined below, with its FORMAT String shown in
parenthesis.
MO (I2) = molecule number
ISO (I1) = isotopologue number (1 = most abundant, 2 = second, etc)
* F (F13.4) = frequency of transition in MHz
* errf (F8.4) = error in f in MHz
S (E10.3) = intensity in cm-1/(molec * cm-2) at 296 K
* AGAM (F5.4) = air-broadened halfwidth (HWHM) in MHz/Torr at Tref
* SGAM (F5.4) = self-broadened halfwidth (HWHM) in MHz/Torr at Tref
E (F10.4) = lower state energy in wavenumbers (cm-1)
N (F4.2) = coefficient of temperature dependence of
air-broadened halfwidth
* N_self (F4.2) = coefficient of temperature dependence of
self-broadened halfwidth
* Tref (F7.2) = reference temperature for AGAM and SGAM
* d (F9.7) = shift of transition due to pressure (MHz/Torr)
V1 (I3) = upper state global quanta index
V2 (I3) = lower state global quanta index
Q1 (A9) = upper state local quanta
Q2 (A9) = lower state local quanta
IERS (I1) = accuracy index for S
IERH (I1) = accuracy index for AGAM
* IERN (I1) = accuracy index for N
The asterisks mark entries that are different from HITRAN.
Note that AGAM and SGAM are for the temperature Tref, while S is
still for 296 K!
The molecule numbers are encoded as shown in the table below:
0= Null 1= H2O 2= CO2 3= O3 4= N2O 5= CO
6= CH4 7= O2 8= NO 9= SO2 10= NO2 11= NH3
12= HNO3 13= OH 14= HF 15= HCl 16= HBr 17= HI
18= ClO 19= OCS 20= H2CO 21= HOCl 22= N2 23= HCN
24=CH3Cl 25= H2O2 26= C2H2 27= C2H6 28= PH3 29= COF2
30= SF6 31= H2S 32=HCOOH 33= HO2 34= O 35= CLONO2
36= NO+ 37= Null 38= Null 39= Null 40=H2O_L 41= Null
42= Null 43= OCLO 44= Null 45= Null 46=BRO 47= Null
48= H2SO4 49=CL2O2
All molecule numbers are from HITRAN, except for species with id's
greater or equals 40, which are not included in HITRAN.
(E.g.: For BrO, iso=1 is Br-79-O,iso=2 is Br-81-O.)
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 31.10.00
Definition at line 1420 of file linerecord.cc.
References convMytranIER(), CREATE_OUT0, CREATE_OUT3, extract(), iso(), SpeciesRecord::Isotopologue(), magam, max, mdagam, mdf, mdi0, mdnair, melow, mf, mi0, misotopologue, mnair, mnself, mpsf, msgam, mspecies, mtgam, mti0, my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, SPEED_OF_LIGHT, TORR2PA, and wavenumber_to_joule().
Referenced by abs_linesReadFromMytran2(), and ARTSCAT4UnusedToNaN().
◆ setAgam()
|
inline |
◆ setF()
|
inline |
Set the line center frequency in Hz.
Definition at line 342 of file linerecord.h.
References mf.
Referenced by abs_lines_per_speciesAddMirrorLines(), and propmat_clearskyAddZeeman().
◆ setI0()
|
inline |
Set Intensity.
Definition at line 365 of file linerecord.h.
References mi0.
Referenced by propmat_clearskyAddZeeman().
◆ setNair()
|
inline |
Set AGAM temperature exponent (dimensionless):
Definition at line 389 of file linerecord.h.
References mnair.
◆ setNself()
|
inline |
Set SGAM temperature exponent (dimensionless):
Definition at line 395 of file linerecord.h.
References mnself.
◆ setPsf()
|
inline |
Set the pressure shift parameter in Hz/Pa.
Definition at line 348 of file linerecord.h.
References mpsf.
◆ setSgam()
|
inline |
◆ Sgam()
|
inline |
Self broadened width in Hz/Pa:
Definition at line 380 of file linerecord.h.
References msgam.
Referenced by calc_gamma_and_deltaf_artscat4(), operator<<(), and xsec_species().
◆ Species()
|
inline |
The index of the molecular species that this line belongs to.
The species data can be accessed by species_data[Species()].
Definition at line 302 of file linerecord.h.
References mspecies.
Referenced by abs_lines_per_speciesCreateFromLines(), ARTSCAT4FromARTSCAT3(), find_matching_lines(), propmat_clearskyAddZeeman(), and xsec_species().
◆ SpeciesData()
| const SpeciesRecord & LineRecord::SpeciesData | ( | ) | const |
The matching SpeciesRecord from species_data.
To get at the species data of a LineRecord lr, you can use:
- species_data[lr.Species()]
- lr.SpeciesData()
The only advantages of the latter are that the notation is slightly nicer and that you don't have to declare the external variable species_data.
Definition at line 50 of file linerecord.cc.
References mspecies, and global_data::species_data.
Referenced by abs_lines_per_speciesCreateFromLines(), and Isotopologue().
◆ Tgam()
|
inline |
Reference temperature for AGAM and SGAM in K:
Definition at line 398 of file linerecord.h.
References mtgam.
Referenced by operator<<(), and xsec_species().
◆ Ti0()
|
inline |
Reference temperature for I0 in K:
Definition at line 368 of file linerecord.h.
References mti0.
Referenced by calc_gamma_and_deltaf_artscat4(), operator<<(), and xsec_species().
◆ Upper_GQuanta()
|
inline |
Broadening parameter self in Hz/Pa :
Broadening parameter N2 in Hz/Pa : Broadening parameter O2 in Hz/Pa : Broadening parameter H2O in Hz/Pa : Broadening parameter CO2 in Hz/Pa : Broadening parameter H2 in Hz/Pa : Broadening parameter He in Hz/Pa : GAM temp. exponent N self: GAM temp. exponent N N2: GAM temp. exponent N O2: GAM temp. exponent N H2O: GAM temp. exponent N CO2: GAM temp. exponent N H2: GAM temp. exponent N He: F Pressure shift N2 in Hz/Pa : F Pressure shift O2 in Hz/Pa : F Pressure shift H2O in Hz/Pa : F Pressure shift CO2 in Hz/Pa : F Pressure shift H2 in Hz/Pa : F Pressure shift He in Hz/Pa : Upper state global quanta
Definition at line 511 of file linerecord.h.
References mupper_gquanta.
◆ Upper_J()
|
inline |
◆ Upper_LQuanta()
|
inline |
◆ Upper_N()
|
inline |
◆ Version()
|
inline |
Return the version number.
Definition at line 298 of file linerecord.h.
References mversion.
Referenced by abs_linesReadFromArtsObsolete(), ARTSCAT4FromARTSCAT3(), operator<<(), xml_write_to_stream(), and xsec_species().
◆ VersionString()
| String LineRecord::VersionString | ( | ) | const |
Return the version String.
Definition at line 34 of file linerecord.cc.
References mversion.
Referenced by abs_linesReadFromArtsObsolete(), LineRecord(), and xml_write_to_stream().
Member Data Documentation
◆ ma
|
private |
Definition at line 990 of file linerecord.h.
Referenced by A(), and ReadFromArtscat4Stream().
◆ magam
|
private |
Definition at line 958 of file linerecord.h.
Referenced by Agam(), ARTSCAT4FromARTSCAT3(), ARTSCAT4UnusedToNaN(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and setAgam().
◆ maux
|
private |
Definition at line 968 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), Aux(), Naux(), and ReadFromArtscat3Stream().
◆ mdagam
|
private |
Definition at line 977 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dAgam(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), and ReadFromMytran2Stream().
◆ mdelta_foreign
|
private |
Definition at line 1040 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), Delta_foreign(), and ReadFromArtscat4Stream().
◆ mdf
|
private |
Definition at line 973 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dF(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), and ReadFromMytran2Stream().
◆ mdi0
|
private |
Definition at line 975 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dI0(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), and ReadFromMytran2Stream().
◆ mdnair
|
private |
Definition at line 981 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dNair(), ReadFromArtscat3Stream(), ReadFromHitran2004Stream(), and ReadFromMytran2Stream().
◆ mdnself
|
private |
Definition at line 983 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dNself(), ReadFromArtscat3Stream(), and ReadFromHitran2004Stream().
◆ mdpsf
|
private |
Definition at line 985 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dPsf(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), and ReadFromHitran2004Stream().
◆ mdsgam
|
private |
Definition at line 979 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), dSgam(), ReadFromArtscat3Stream(), and ReadFromHitran2004Stream().
◆ melow
|
private |
Definition at line 956 of file linerecord.h.
Referenced by Elow(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), and ReadFromMytran2Stream().
◆ mf
|
private |
Definition at line 948 of file linerecord.h.
Referenced by F(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and setF().
◆ mgamma_foreign
|
private |
Definition at line 1002 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), Gamma_foreign(), and ReadFromArtscat4Stream().
◆ mglower
|
private |
Definition at line 994 of file linerecord.h.
Referenced by G_lower(), and ReadFromArtscat4Stream().
◆ mgupper
|
private |
Definition at line 992 of file linerecord.h.
Referenced by G_upper(), and ReadFromArtscat4Stream().
◆ mi0
|
private |
Definition at line 952 of file linerecord.h.
Referenced by I0(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and setI0().
◆ misotopologue
|
private |
Definition at line 946 of file linerecord.h.
Referenced by Isotopologue(), IsotopologueData(), Name(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), and ReadFromMytran2Stream().
◆ mlower_gquanta
|
private |
Lower state global quanta.
Definition at line 1058 of file linerecord.h.
Referenced by Lower_GQuanta(), and ReadFromHitran2004Stream().
◆ mlower_j
|
private |
Lower state local J quanta.
Definition at line 1070 of file linerecord.h.
Referenced by Lower_J(), ReadFromArtscat4Stream(), and ReadFromHitran2004Stream().
◆ mlower_lquanta
|
private |
Lower state local quanta.
Definition at line 1062 of file linerecord.h.
Referenced by Lower_LQuanta(), and ReadFromHitran2004Stream().
◆ mlower_n
|
private |
Lower state local N quanta.
Definition at line 1068 of file linerecord.h.
Referenced by Lower_N(), ReadFromArtscat4Stream(), and ReadFromHitran2004Stream().
◆ mn_foreign
|
private |
Definition at line 1023 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), N_foreign(), and ReadFromArtscat4Stream().
◆ mnair
|
private |
Definition at line 962 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), ARTSCAT4UnusedToNaN(), Nair(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and setNair().
◆ mnself
|
private |
Definition at line 964 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), Nself(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and setNself().
◆ mpsf
|
private |
Definition at line 950 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), ARTSCAT4UnusedToNaN(), Psf(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and setPsf().
◆ mquantum_numbers
|
private |
Quantum numbers from HITRAN.
Definition at line 1076 of file linerecord.h.
Referenced by QuantumNumbers(), ReadFromArtscat4Stream(), and ReadFromHitran2004Stream().
◆ mquantum_numbers_str
|
private |
String with quantum numbers for ARTSCAT-4.
Definition at line 1073 of file linerecord.h.
Referenced by QuantumNumbersString(), and ReadFromArtscat4Stream().
◆ msgam
|
private |
Definition at line 960 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), setSgam(), and Sgam().
◆ mspecies
|
private |
Definition at line 944 of file linerecord.h.
Referenced by IsotopologueData(), Name(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), Species(), and SpeciesData().
◆ mtgam
|
private |
Definition at line 966 of file linerecord.h.
Referenced by ARTSCAT4UnusedToNaN(), ReadFromArtscat3Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and Tgam().
◆ mti0
|
private |
Definition at line 954 of file linerecord.h.
Referenced by ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromJplStream(), ReadFromMytran2Stream(), and Ti0().
◆ mupper_gquanta
|
private |
Upper state global quanta.
Definition at line 1056 of file linerecord.h.
Referenced by ReadFromHitran2004Stream(), and Upper_GQuanta().
◆ mupper_j
|
private |
Upper state local J quanta.
Definition at line 1066 of file linerecord.h.
Referenced by ReadFromArtscat4Stream(), ReadFromHitran2004Stream(), and Upper_J().
◆ mupper_lquanta
|
private |
Upper state local quanta.
Definition at line 1060 of file linerecord.h.
Referenced by ReadFromHitran2004Stream(), and Upper_LQuanta().
◆ mupper_n
|
private |
Upper state local N quanta.
Definition at line 1064 of file linerecord.h.
Referenced by ReadFromArtscat4Stream(), ReadFromHitran2004Stream(), and Upper_N().
◆ mversion
|
private |
Definition at line 942 of file linerecord.h.
Referenced by ARTSCAT4FromARTSCAT3(), ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), Version(), and VersionString().
The documentation for this class was generated from the following files:
